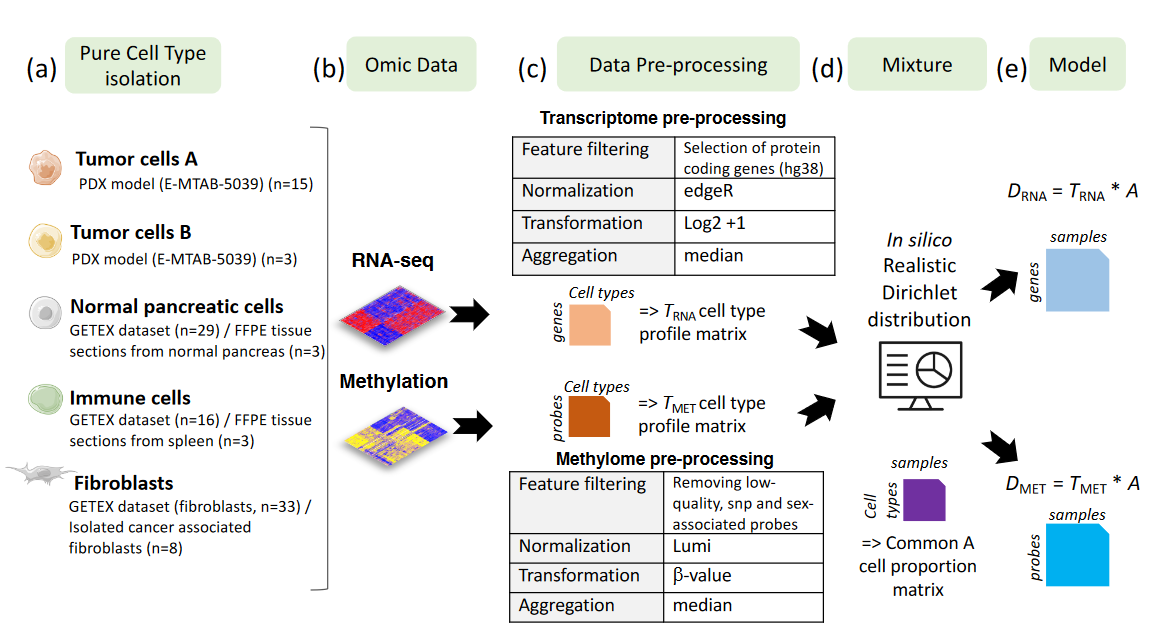
- 5 different cell populations present in pancreatic tumors were considered.
- Raw transcriptome and methylome profiles of these different cell populations were extracted from various sources (PDX model, tissues or isolated cells).
- Raw cell type profile matrices were preprocessed (Feature filtering, normalization, signal transformation, sample aggregation) to avoid any batch effect.
- In silico Dirichlet distribution have been used based on realistic proportions defined by the anatomopathologist expertise (J. Cros).
- Paired methylome and transcriptome of in silico mixtures from pancreatic tumors were obtained by considering D = T A, with T the cell-type profiles (matrix of size M * K, with M the number of features and K=5 the number of cell types) and A the cell-type proportion per patient (matrix of size K * N, with N=30 the number of samples) common between both omics.
More details in our article